The Metabolomics Integrator (MINT) is a web-based post-processing tool for liquid chromatography-mass spectrometry (LC-MS) based metabolomics. MINT enables researchers to extract, visualize, and analyze targeted metabolomics data with an intuitive browser-based interface.
Metabolomics is the comprehensive study of small molecule metabolites in biological samples, such as bacterial cultures or human blood. These metabolites serve as critical biomarkers for:
- Disease diagnostics and treatment development
- Pathogen identification (e.g., methicillin-resistant Staphylococcus aureus [MRSA])
- Drug discovery and pharmacology
- Environmental and agricultural research
MINT streamlines the processing of LC-MS data by providing:
- Targeted peak extraction with automated and manual retention time optimization
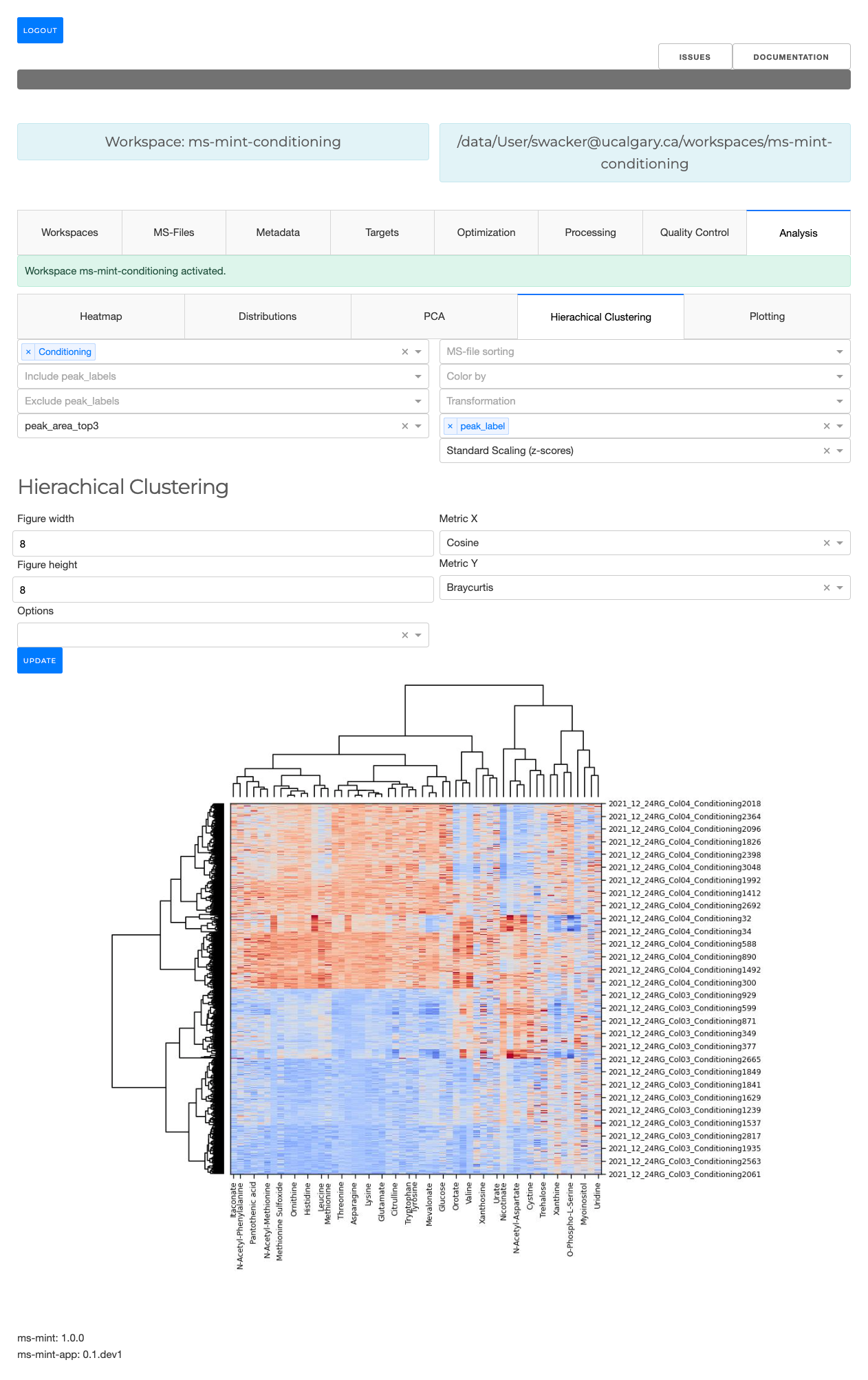
- Interactive visualizations including heatmaps, PCA, and hierarchical clustering
- Quality control tools for assessing data integrity
- Flexible data export for downstream statistical analysis
- Plugin architecture for extending functionality
- Installation Guide - Install MINT via pip, conda, Docker, or Windows installer
- Quickstart Tutorial - Step-by-step guide with demo data
- Demo Data - Download test data with sample LC-MS files and target lists
- Full Documentation - Comprehensive user guide and reference
- Import LC-MS data in mzML and mzXML formats
- Define targeted extraction protocols with customizable m/z and retention time windows
- Automated peak detection and retention time optimization
- Batch processing for large datasets
- Real-time peak shape visualization and quality assessment
- Principal Component Analysis (PCA) for dimensionality reduction
- Hierarchical clustering with dendrograms
- Customizable heatmaps with correlation analysis
- Statistical visualizations (box plots, violin plots, distributions)
- Mass accuracy (m/z drift) monitoring
- Peak shape evaluation across samples
- Sample type categorization (biological, QC, standards)
- Batch effect visualization
- Workspace-based project organization
- Comprehensive metadata integration
- Multiple export formats (CSV, Excel)
- Support for large-scale studies
-
Brown K, Thomson CA, Wacker S, et al. Microbiota alters the metabolome in an age- and sex-dependent manner in mice. Nat Commun. 2023;14:1348.
-
Ponce LF, Bishop SL, Wacker S, Groves RA, Lewis IA. SCALiR: A Web Application for Automating Absolute Quantification of Mass Spectrometry-Based Metabolomics Data. Anal Chem. 2024;96:6566–6574.
MINT is developed by the Lewis Research Group. For other metabolomics and computational biology tools, visit lewisresearchgroup.org/software.
This project uses the ms-mint conda environment. For development and contributing, please refer to the Installation Guide for setting up the conda environment.
We welcome contributions of all kinds! Whether you're reporting bugs, suggesting features, improving documentation, or contributing code, your help makes MINT better for everyone.
- Report Issues: GitHub Issues
- Contribution Guidelines: See CONTRIBUTING.md for development setup and workflow
- Code Standards: This project follows PEP 8 and uses Black and Flake8 for code formatting
If you use MINT in your research, please cite:
Each release is assigned a DOI for proper citation. Visit the DOI link above for version-specific citations.
MINT is built on the shoulders of the open-source community. We are grateful for:
- The Python scientific computing ecosystem (NumPy, Pandas, SciPy, Scikit-learn)
- Plotly Dash for the interactive web framework
- GitHub, Docker Hub, and PyPI for hosting and distribution
- The Plotly Community and Stack Overflow for invaluable support
Special thanks to contributors:
- @rokm - Refactored PyInstaller spec for Windows packaging
- @bucknerns - Versioneer configuration
We also thank all users and early adopters whose feedback and bug reports drive continuous improvement.
MINT is licensed under the Apache License 2.0.